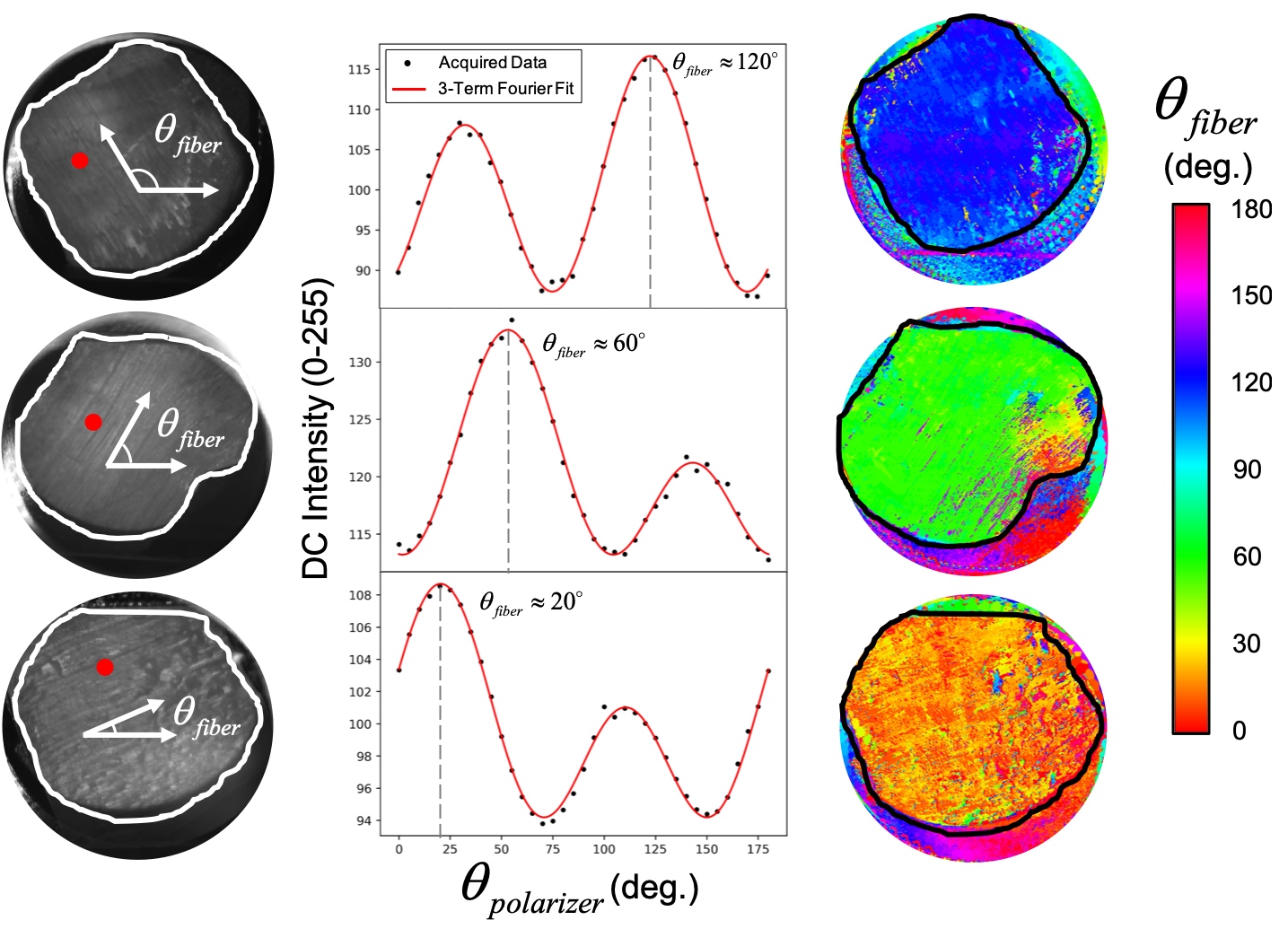
The mitral and tricuspid heart valves are composed of leaflets, or thin flaps of membranous tissue, anchored proximally to a fibrous ring known as the valve annulus and distally to a series of chordae tendineae. These leaflets coapt, or come together, during systole to prevent the passage of blood. During diastole, the leaflets recede into the ventricles to allow passage of blood. This leaflet motion is repeated each heartbeat, leading to profound cyclic loading and cyclic strain on the valve tissue. To handle the imposed loading, the heart valve leaflets are composed of a fibrous network of collagen fibers. These fibers are responsible for the anisotropy, or directional-dependence, of the leaflet tissue’s mechanical behavior. Briefly, the local orientations of the fiber dictate the direction of maximum stiffness in the heart valve leaflets. However, part of the versatility and utility of the collagen fibers in bearing load arises from the fibers’ ability to realign to bear the necessary load. That is to say, the orientations of the fibers are likely to change under applied loading. Although previous groups have examined the collagen fibers in an unloaded state, no prior studies have examined and quantified how these local fiber orientations can change under applied loading. Our study seeks to quantify the changes in local orientations of collagen fibers in heart valve leaflets under dynamic applied loading, and to present this data via colormaps like the one shown above.
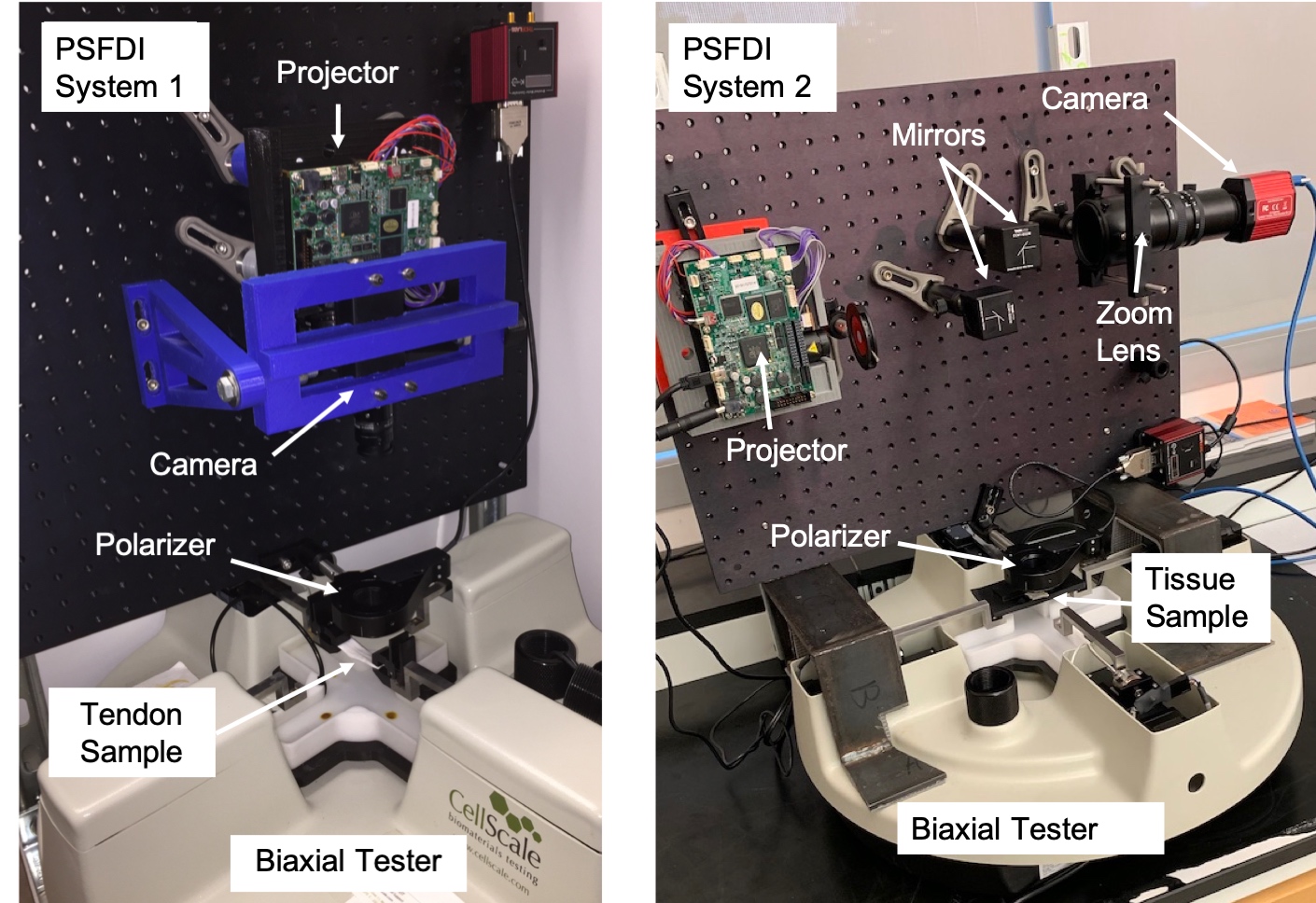
To examine these collagen fibers, our group built a system reliant upon a novel technique known as “polarized Spatial Frequency Domain Imaging”. This technique capitalizes on the birefringence, or polarization-dependent, refractive response of the collagen fibers. To capture the birefringence, our system uses a projector, a polarizer, and a 5 MegaPixel CCD camera to capture high resolution images of the testing sample at various polarization angles. We then analyze the images and compute a model fit comparing the intensity recorded at each pixel to the polarization angle. From this fit, we are able to discern the local collagen fiber orientation. These predicted orientations can then be plotted on colormaps like the ones shown above. Our collagen fiber predictions for bovine tendon samples, with spatially homogeneous collagen structures, are shown below.
This project has been a foray into imaging systems and has afforded me a glimpse into the complexity and depth of the scientific imaging field. Through research on advanced imaging techniques like spatial frequency domain imaging, rasterized light-scattering, and polarization microscopy, I’ve realized how much data can be gained about a tissue or biomaterial by simply “looking at it” in an appropriate way. Our data analysis pipeline was implemented in Python, with support from open-source software like GitHub, Anaconda, NumPy, Matplotlib, and many other libraries for Python. I developed the testing scripts for the system through LabView using USB communication protocol to send signals to our projector. Through developing the pipeline, I also learned to refactor code into discrete functions to make a more robust and clearer program. We expect to publish an article about our system and the data obtained. For more information and to see a poster about the project, I’ve attached a recent presentation below.